Cytogenetics Tool
The “Cytogenetics Tool” offers the search for currently available data on genome size, chromosome numbers, and ploidal level, and currently ca. 14% and 55%, respectively, out of the 3977 species from the Brassicaceae are incorporated in BrassiBase for genome size and chromosome numbers, respectively. Taxonomic groups from genera and species down to subspecies are searchable.
The phylogenetic backbone for the Cytogenetics Tool” is based on a synpopis of published comprehensive phylogenetic analyses.
The search field accepts scientific plant names, i. e. names of genera or Brassicaceae tribes.
Examples:
- Arabideae — will find information about all member genera of tribe Arabideae.
- Arabis — will find information about the genus Arabis.
- Arabis alpina — will limit the search to the species Arabis alpina
After having typed 3 characters, you'll be provided with suggestions from the database.
Registered users can also use the advanced search to find data by combinations of values. Choose fields of interest and set criteria using dropdown menus. Fill in desired values (numbers or Y/N).
Genome size data and chromosome numbers are fundamental information to characterize genomes, which are very dynamic in plants in both size and structure. Genome sizes in the Brassicaceae family are comparatively small on average (1C = 0.51 pg). Only a few Brassicaceae species from various tribes of Lineage III reach genome sizes up to 1C = 4.31 pg. Genome up- and downsizing has been shown to be rapid in the Brassicaceae family (Lysak et al., 2009), and the number of polyploids is with about 65% in congruence with the 50-70% portion of polyploids estimated for all extant angiosperms (Wendel, 2000). Tetraploidy is most frequent within the Brassicaceae, and numerous genera are characterized by high-polyploidy (e.g., Draba). Taxa frequently occur both as diploid and polyploid cytotypes.
Genome sizes were obtained from literature; chromosome counts were assembled from Warwick et al. (2006), and additional counts are ongoingly added from the literature since 2006. Ploidal levels were estimated based on literature and in collaboration with Martin Lysak (Brno). For all datasets, literature values were subjected to a critical quality check, and the newest taxonomic concept was applied.
- Hohmann, H, Wolf, E, Lysak, M, Koch MA. 2015. A Time-Calibrated Road Map of Brassicaceae Species Radiation and Evolutionary History. The Plant Cell 27: 2770-2784.
- Lysak, M.A., Koch, M.A., Leitch, I.J., Beaulieau, J.M., and Meister, A. (2009). The dynamic ups and downs of genome size evolution in Brassicaceae. Mol. Biol. Evol. 26: 85–98.
- Warwick, S.I. and Al-Shehbaz, I.A. (2006). Brassicaceae: Chromosome number index and database on CD-Rom. Plant Syst. Evol. 259: 237–248.
- Wendel J. F. 2000. Genome evolution in polyploids. Plant Molecular Biology 42: 225-249.
Cytogenetic Data Plots
Standard phylogenetic diagram with genome sizes  | Standard phylogenetic diagram with cytogenetic information  | Average genome sizes within tribes 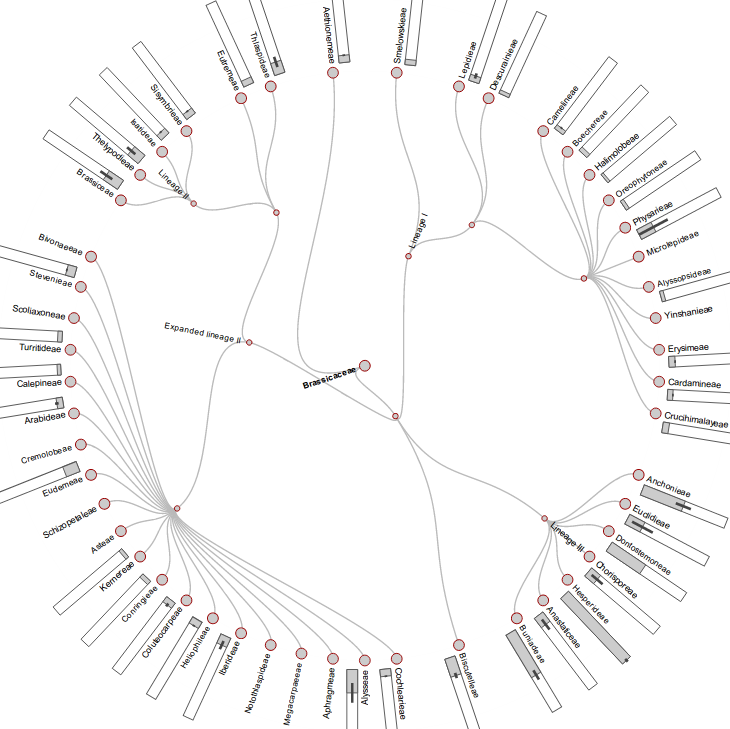 |